
 Jun Li
Jun Li
Notre Dame researchers representing four labs across two colleges at the University have received a four-year, $1.1 million Research Project Grant (R01) from the National Institute of General Medical Sciences at the National Institutes of Health (NIH). The oldest grant mechanism used by the NIH, the R01 provides support for health-related research and development based on the mission of the NIH.
 Patricia Clark
Patricia Clark
Principal Investigators of this grant include two from the College of Science: Jun Li, Ph.D. of the department of Applied and Computational Mathematics and Statistics (ACMS) and Patricia Clark, Ph.D. of the department of Chemistry and Biochemistry; and two from the College of Engineering: Scott Emrich, Ph.D. and Tijana Milenkovic, Ph.D. both of the department of Computer Science and Engineering.
The awarded project, titled “Integrative Computational Framework for Pattern Mining in Big -omics Data: Linking Synonymous Codon Usage to Protein Biogenesis,” expands upon a line of inquiry started several years ago by Clark and Emrich, who sought to develop a computational approach to test the hypothesis that small changes to the rate of protein synthesis could change the folding of the encoded protein.
Efficient production of proteins is arguably the most important function of a cell. In the cell, proteins are synthesized as linear polymers, but must fold into a three-dimensional shape in order to function. Sometimes, the rate of folding can be faster than the rate of synthesis, which can lead to diseases like Alzheimer’s disease, cystic fibrosis, diabetes, liver diseases, blood clotting or bleeding disorders, or even cancer, among other diseases.
 Tijana Milenkovic
Tijana Milenkovic
Early during the development of this project, Clark and Emrich began collaborating with Li and Milenkovic: Li for his expertise in statistical modeling and mining on big data and Milenkovic for her expertise in network science. In summary, what the researchers found was that clusters of slow codons do tend to co-occur at similar positions in genes encoding proteins with similar structures, and this co-occurrence is both widespread and statistically significant. But why certain genes have these co-occurring clusters was much harder to figure out, and forms the basis for the new R01 award.
 Scott Emrich
Scott Emrich
Emrich and Milenkovic are developing new sequence and network analysis methods, respectively, made possible by Li’s novel statistical framework for efficient mining of interesting patterns in large-scale genomic/proteomic sequence and structural network data. Critical to the validity of computational analysis, and to make the experimental validation cost-efficient, Li will also develop rigorous statistical tests to rule out possible false positive discoveries and recommend the most promising patterns and hypotheses for further experimental validation. Clark’s lab is working on the computational analyses with other investigators and developing experiments in the lab to construct gene sequences with different patterns of synonymous codon usage and test hypotheses developed from their computational and statistical approaches. Results from the experimental studies will be used to construct an iterative feedback loop between the computational/statistical results and hypotheses regarding the specific roles for rare codon clusters in efficient protein production.
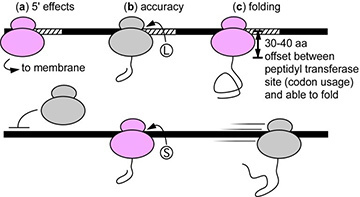 Figure 1. Examples of potential effects of synonymous rare codons (hatched areas) on protein biogenesis. Black bar = mRNA; pink and grey ovals = ribosomes. Note that in some cases rare codons are expected to improve biogenesis (pink ribosomes), whereas in other cases rare codons will impede biogenesis (grey ribosomes). These impacts include: (a) 5’ rare codons that improve translation initiation efficiency 12 or membrane targeting 9; (b) rare codons that increase mistranslation (leading to the incorrect incorporation of an amino acid, i.e., L vs. S) 22; © rare codons that retard translation rate to improve co-translational folding of the encoded protein [11, 14-16]; and (d) other effects (not shown): synonymous substitutions have also been shown to affect mRNA degradation rate 23, splicing, and post-translational modifications 24.
Figure 1. Examples of potential effects of synonymous rare codons (hatched areas) on protein biogenesis. Black bar = mRNA; pink and grey ovals = ribosomes. Note that in some cases rare codons are expected to improve biogenesis (pink ribosomes), whereas in other cases rare codons will impede biogenesis (grey ribosomes). These impacts include: (a) 5’ rare codons that improve translation initiation efficiency 12 or membrane targeting 9; (b) rare codons that increase mistranslation (leading to the incorrect incorporation of an amino acid, i.e., L vs. S) 22; © rare codons that retard translation rate to improve co-translational folding of the encoded protein [11, 14-16]; and (d) other effects (not shown): synonymous substitutions have also been shown to affect mRNA degradation rate 23, splicing, and post-translational modifications 24.
Originally published by Tammi Freehling at science.nd.edu on September 06, 2016.